Scientists and companies invest years and fortunes searching for new medicines. Many discoveries might already exist but remain hidden in the vast scientific literature. Ramakrishnan Kannan, group leader for discrete algorithms at Oak Ridge National Laboratory, develops tools to mine that gigantic and complex information space.
Kannan’s work elevates an existing research field. In 1986, the late Don Swanson, a University of Chicago humanities professor, described “undiscovered public knowledge.” By searching the scientific literature, he found that fish oil might treat an autoimmune condition, Reynaud’s disease. A couple years later, Swanson used a similar approach to recommend magnesium to treat migraines.
“Swanson just read through the bunch of publications and started suggesting medicines for people with diseases,” says Kannan, who collaborates with Thomas Potok, ORNL; Rich Vuduc, Georgia Institute of Technology; and Sergio Baranzini, University of California San Francisco. “Around the 2000s, this became popularly known as literature-based discoveries, and it’s a billion-dollar industry nowadays.”
Mining publications for medical knowledge depends on making connections, Kannan explains. “It’s a very complex relation that you have to capture from the publications – for example, starting with human biology, genomics, and their interconnections, like epigenetics, protein expression, tissues and organs, diseases and conditions.”
Swanson’s manual approach would fail in the face of today’s 35 million papers. “Plus, there are about 360,000 medical concepts in those papers,” Kannan says. “That creates about 10 billion connections to analyze, which is humanly impossible.”
Instead of tediously and endlessly flipping through journal articles, scientists need a formal approach to find relationships between medical concepts, based on results obtained in specific experiments. Today, this field is known as biomedical knowledge graphs, or KGs.
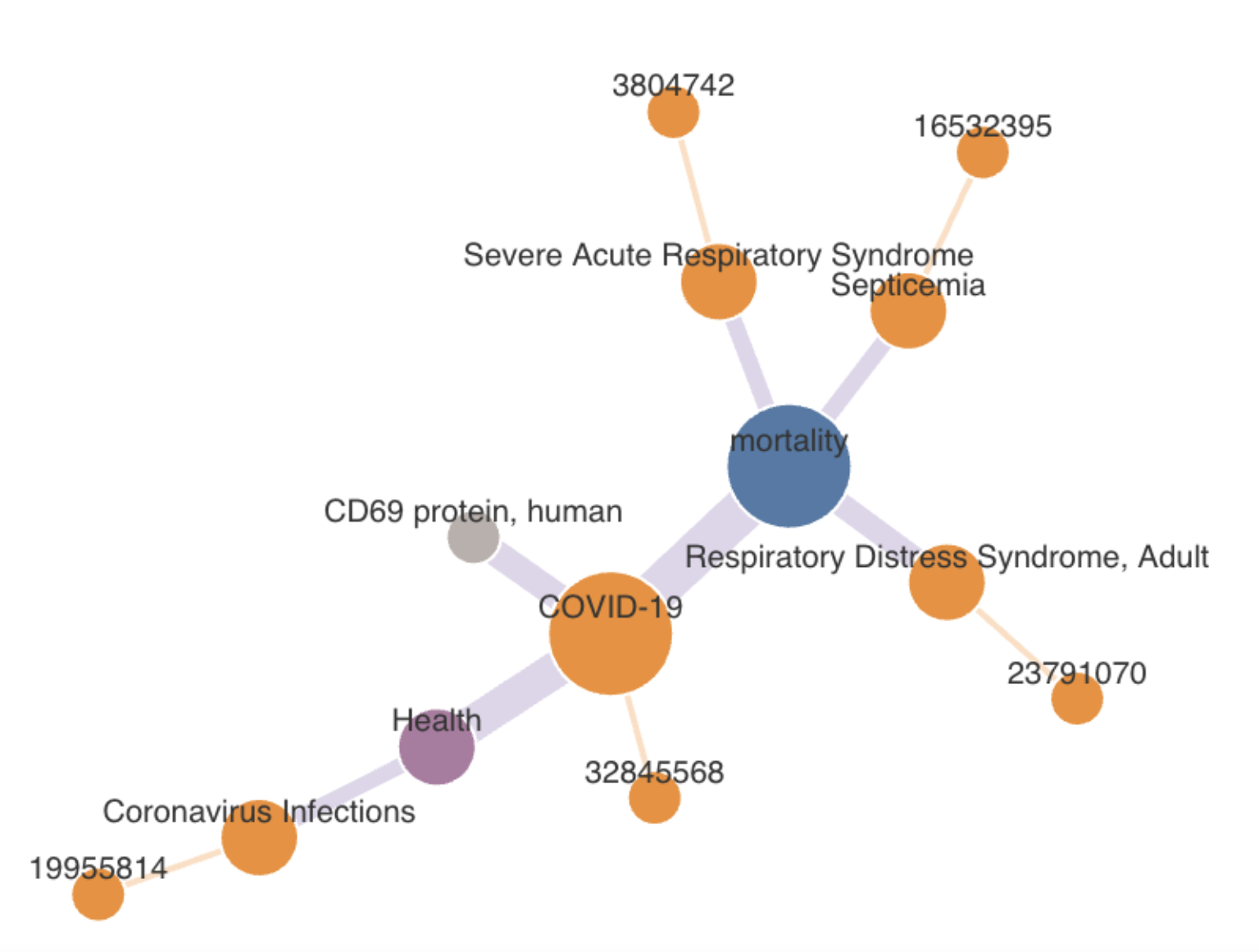
Based on information in annotated-text databases such as PubMed, KGs represent connections between medical concepts, which can be analyzed with an all-pairs shortest-paths algorithm – basically showing which ideas are the most closely connected. Then, the connection between concepts can be compared with curated biomedical KGs, such as UCSF’s SPOKE. New connections found by computer-driven KGs can be especially useful, Kannan says, but it’s not clear why curated KGs miss some important connections. “We want to understand that gap “between known and undiscovered relationships.
‘It takes four and a half years for (all of) humanity to perform what we can do in a single second on Frontier.’
To analyze the billions of relationships between hundreds of millions of concepts in tens of millions of articles, Kannan and his team, working with processor company AMD, developed COAST (exascale communication-optimized all-pairs shortest path), a graph-based artificial intelligence. Kannan’s team explored COAST on a subset of 35 million PubMed papers.
But creating and analyzing KGs fast enough for biomedical industry use requires a GPU-accelerated supercomputer, such as the Oak Ridge Leadership Computing Facility’s Frontier. To explain why graphics processing units, which work with standard processors to accelerate calculations, are needed, Kannan offers this comparison: “If you give a calculator to the 8 billion people in the world and ask them to perform one multiplication every second, it takes four and a half years for (all of) humanity to perform what we can do in a single second on Frontier.”
Kannan’s team moved onto Frontier slowly. “To understand the COAST algorithm, we first ran it on a small set of nodes,” he says. “Once we understand that, we want to run it on Frontier’s 75,000-plus GPU GCDs,” or graphics compute dies. Each of the more than 9,000 Frontier nodes has four AMD GPUs, and each GPU has two GCDs. As a result, COAST became the first graph-AI applications to run at more than one exaflop, which is a quintillion calculations per second.
The scientists have found performance variations while scaling up the code. Although using more nodes seems like it would increase performance, that doesn’t always happen. With a graph of performance versus the number of nodes, a polynomial fit to the data – essentially tracking the trends in the data with a curved lined – predicts what should happen with more nodes. “This is called hyperbolic performance modeling,” Kannan says. If there is a gap between extrapolating the polynomial and experimental findings, “we have to understand whether that is due to the system or that is due to the algorithm.” The team came up with HYPERMOD (hyperbolic performance models) “to understand the performance of our algorithm from a few dozen GPUs to a few tens of thousands of GPUs on Frontier,” Kannan explains.
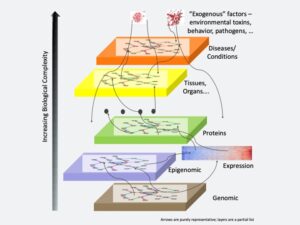
The scientific literature contains valuable knowledge for developing new medicines but finding it creates a huge challenge. The diverse levels of information—from genomics to diseases—must be connected, despite the increasing biological complexity. Image courtesy of Ramakrishnan Kannan.
To employ COAST and HYPERMOD, Kannan and his colleagues compared biomedical information in journals and KGs. “We tried to understand the relations between results in publications against a hand-curated knowledge graph like SPOKE,” Kannan says. In this work, the scientists created three relation classes: a match, a partial match, and results that exist in only one database.
That work showed that about 40% of the relations found in SPOKE are not in PubMed. “SPOKE includes discoveries that have never been tagged in the community,” Kannan says. “That means there are still a lot of things to be done.”
When considering the cause of that knowledge gap, Kannan factors in analysis, time, and money required to write a paper. Thus, some biomedical connections will be missed, he says. “People are not paying attention or it is not their priority, and tools like COAST can help the scientists understand their toenail-size research contribution toward understanding an unknown gigantic dinosaur like COVID.”
Nonetheless, the gap appears to be closing. In a study of a 2020 publication dataset, Kannan’s team found that the research community is starting to discuss more paths that are available in SPOKE. Some of the paths that appear also have gotten shorter, meaning tighter connections are forming between related data.
To help more scientists use biomedical KGs, Kannan and his colleagues have open-sourced some of their algorithms.
He and his partners hope that information their KGs unearth will become more widely available. “We want to build our confidence in links discovered with algorithms like COAST to see if we can add a discovery aid that can be listed on other repositories, like SPOKE.”
COAST’s capabilities, though, could go beyond biomedicine. “We want to replicate the success that we observed in biomedical knowledge graphs to other domains, like chemistry, materials, and physics,” Kannan says.
In that way, KGs might reach Swanson’s broad idea of finding undiscovered public knowledge. One day, KGs could become a go-to tool for mining information that changes many aspects of the world’s technology and people’s everyday lives.