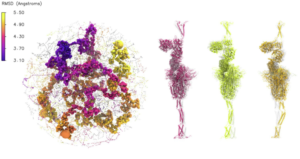 Researchers have used computational screens that incorporate molecular dynamics and machine learning mostly for drug discovery, but these techniques also have played a key role in understanding the SARS-CoV-2 virus (left). The project took the DeepDriveMD software in new directions. The team collaborated with Rommie Amaro of the University of California, San Diego, and her colleagues on dramatic work that simulated the virus’ exterior spike protein interacting with the human ACE2-receptor protein. The full envelope consists of more than 300 million atoms, including proteins and an extensive network of carbohydrates – sugar molecules – that envelop the virus surface. The three structures to the right of the whole virus depict the spike protein with different motions in the stalk region, the helices at the bottom of each. The colors (magenta, yellow and orange) correspond with the regions in the left image.
Researchers have used computational screens that incorporate molecular dynamics and machine learning mostly for drug discovery, but these techniques also have played a key role in understanding the SARS-CoV-2 virus (left). The project took the DeepDriveMD software in new directions. The team collaborated with Rommie Amaro of the University of California, San Diego, and her colleagues on dramatic work that simulated the virus’ exterior spike protein interacting with the human ACE2-receptor protein. The full envelope consists of more than 300 million atoms, including proteins and an extensive network of carbohydrates – sugar molecules – that envelop the virus surface. The three structures to the right of the whole virus depict the spike protein with different motions in the stalk region, the helices at the bottom of each. The colors (magenta, yellow and orange) correspond with the regions in the left image.
At first, even using all of the Summit supercomputer’s considerable computational resources, the team could sample spike protein-ACE2-receptor interaction for only a paltry 500 nanoseconds, too fleeting for a meaningful model. So the DeepDriveMD team sped up the simulation, increasing the numbers of protein conformations they could sample by more than 8-fold. The project won the 2020 Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research. The overall understanding of the virus it provides has informed drug and vaccine development.
Image courtesy of Anda Trifan, John Stone, Lorenzo Casalino, Alexander Brace, Arvind Ramanathan and Rommie Amaro.