For decades, scientists have probed how human DNA that would stretch more than 2 meters if unspooled from a cell is packed into chromosomes millions of times smaller.
What’s more, they’ve tried to learn how this extreme folding influences genes. They way genes are packed into chromosomes can produce cells that smoothly develop into organs and bones or that misfire and lead to disease.
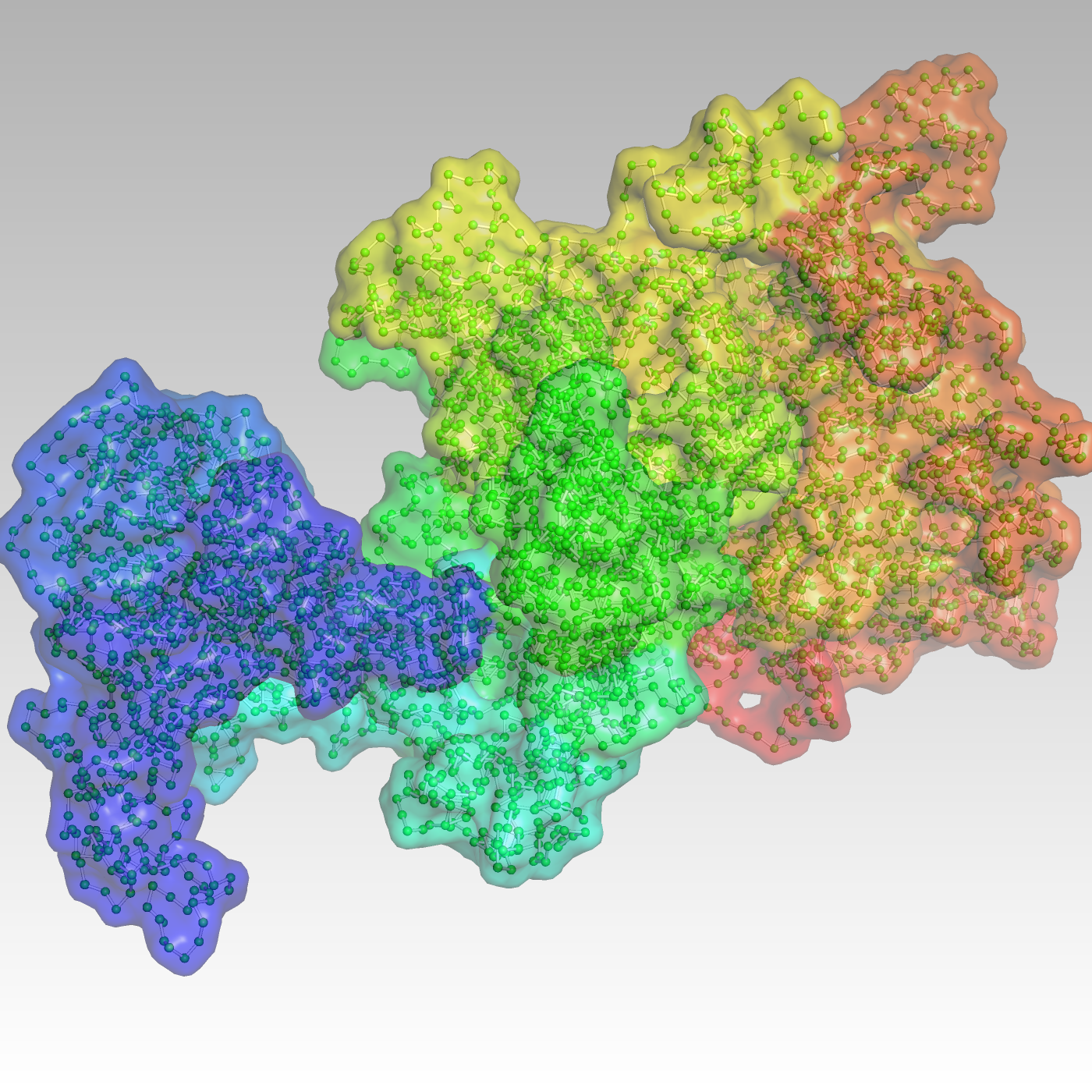
But with current techniques to investigate folding, “we’re chasing shadows of the reality,” says Jie Liang, a professor in the Richard and Loan Hill biomedical engineering department at the University of Illinois Chicago (UIC), where he leads the Molecular and Systems Computational Bioengineering Lab. Researchers require detailed, physics-based, three-dimensional models of these important structures to understand their critical cellular machinery.
Liang and his colleagues say they have an answer: an algorithm that produces just such models. The team is extending its capabilities with time on Argonne National Laboratory supercomputers, thanks to a 2023 INCITE (Innovative and Novel Computational Impact on Theory and Experiment) grant from the Department of Energy’s Advanced Scientific Computing Research program.
It takes computer power to unsnarl the gnarly nature of a genome, the genes packed into our cells. To pack so much DNA into such a tiny spaces, the genes are first wound around proteins called histones, then folded into chromatin, the intricate structures that comprise chromosomes.
Chromatin folding makes neighbors of genes that would be distant on a stretched DNA chain. That influences how they’re expressed because nearby genes in 3D often work together. Knowing which genes are near one another can help reveal factors driving expression, including how cells differentiate into skin, bone, muscle and organs. Liang’s co-investigator, Konstantinos Chronis, a biochemistry and molecular genetics professor in the university’s medical college, focuses on this, seeking ways to reprogram cells so they grow into particular tissues. His research could lead to lab-grown flesh and bone.
“It’s basically physical control of a factory – what gets made, what gets shut down,” Liang says. Misfolded chromatin can produce cells that “are not doing the right job, or they go crazy, and you might have cancer.”
Researchers use a process called chromosome conformation capture to study genome organization. The technique, particularly a version named Hi-C, produces two-dimensional heat maps of the probabilities that genes are near each other. The method, however, averages responses from multiple cells to find these associations, and studies of single cells often find their chromosome structures vary widely. Other technology captures high-resolution images of folding in single cells but only for a limited number across a small area.
Hi-C and similar technology are useful for biology and genomics research, Liang says, but produce just “a glimpse of the shadow projected on 2D.”
Noise in the heat map data also can obscure results. Folding puts many chromatin strands near each other, but that doesn’t necessarily mean their genes link. “You are seeing all of that – signals, noise together” in Hi-C data. “It’s very difficult to sort this out without computational modeling.”
The repetitious random testing of potential folds demands computer power.
The approach Liang, Chronis and colleagues use is based on CHROMATIX, a method whose chief developer was former graduate student Alan Perez-Rathke. It first creates an ensemble of random chromatin structures. The algorithm computes the statistical significance of interactions in Hi-C data by comparing them with the ensemble. It removes unimportant interactions and uses the remaining valid ones as constraints to build 3D chromatin models, at varying resolutions, for a genome region or an entire chromosome.
Building the 3D ensemble is a Bayesian process, with the code repeatedly constructing and sampling chromatin conformations. The repetitious random testing of potential folds demands computer power, Liang says, and the physics governing their behavior makes it even more difficult. He notes, for example, that chromatin strands avoid crossing even as they’re packed into a microscopic space.
Imagine, Liang says, standing in a small room in which you must walk 2,000 steps in a line “and you cannot step on wherever you have already stepped; you have to be self-avoiding.” Finding these patterns in chromatin requires testing myriad options. Even with smart algorithms, “this large-scale computation, without the DOE resources, is not possible.”
The algorithm measures distance frequencies between configurations to help identify significant structural relationships. Results are integrated with other information, such as studies of gene promoter activity, to reveal additional biological information. The team checks its results against Hi-C and imaging data. Collaborators will tap the team’s result to guide lab experiments.
Liang and his colleagues have used UIC clusters and the university’s Extreme high-performance computer to calculate conformations for up to 50 loci, or regions where chromatin strands interact – but it consumed months of continuous computing. That’s changed with the team’s INCITE allocation of 1.5 million node-hours on Theta, a Cray XC40, and 125,000 node-hours on Polaris, an HPE Apollo 6500, both at the Argonne Leadership Computing Facility. With help from Argonne experts, the team has rewritten the code to run on graphics processing units, computer chips that accelerate calculations. It’s running 10 times faster on Polaris than it did on machines using only conventional processors.
“The DOE INCITE grant is really fantastic,” he says, because it provides the power needed to compute up to 3,000 loci in each of three or four types of human cells. “This is unprecedented.”
Liang and the team have focused their early efforts on identifying where chromatin folding itself happens. Revealing that, he says, can help decipher interactions that control how other genes are expressed. In papers describing chromatin folding in fruit flies and human cells, they report that only a tiny portion of interactions – from 10% to as little as 1% or less – guide the process. It’s “shocking news,” Liang says.
“These are the really important interactions that are likely driving chromatin folding,” he says. “One of the things we hope to do with the Argonne resources is to really figure out what is driving (folding) and what are the key physical interactions.”
The team also found that patterns change depending on an organism’s developmental stage. “Certain types of interactions gradually reduce, and certain types gradually increase,” Liang says – variations that are undetectable in Hi-C data.
Modeling also can help measure cell heterogeneity, as they differentiate into skin, bone or other tissues. General patterns drive the process across all organisms and cells, but each one also is different. “How can you get a handle of this heterogeneity? With these models we can really build a quantitative measurement.”
Liang hopes the supercomputing time will lead to a human chromatin atlas that researchers could use to identify gene structures at specific chromosome locations and learn how they control expression.
“We really hope, maybe writing another INCITE proposal, to get the whole genome everywhere” at high resolution, he says. “We also want to do it in coarse grain resolution, where you look at all these, say, 46 chromosomes together.”