In the old days of cell biology, scientists knew the atomic structures of few membrane proteins. Now, amid the so-called resolution revolution brought about by super-precise microscope technology, scientists witness novel structures of new cell membrane proteins almost every week.
“There are always mechanistic questions about these proteins,” says Emad Tajkhorshid, J. Woodland Hastings Endowed Chair in Biochemistry at the University of Illinois at Urbana-Champaign. “There are opportunities that we are not tapping into because we don’t have a complete view of the structure and dynamics of viral particles.”
Tajkhorshid plans to change all that for the insect-borne Zika virus, among others. “The hope is that we will be able to identify processes or behaviors of this viral particle that can be used as targets in pharmaceutical companies to develop drugs,” he says.
Tajkhorshid and his team have used Titan, a now-retired Cray XK7 supercomputer at the Department of Energy’s Oak Ridge National Laboratory, to run simulations on important viral processes with nanoscale molecular dynamics (NAMD) software. But soon, the team will run NAMD on Frontier, the nation’s first exascale supercomputer, roughly 50 times faster than today’s top high-performance systems.
NAMD is one of eight research codes that the Oak Ridge Leadership Computing Facility (OLCF) selected to take part in the Frontier Center for Accelerated Readiness (CAAR) program. CAAR projects ran on OLCF’s precursor systems with Frontier’s early-generation hardware platforms to ensure a smooth transition for the new machine’s users in 2022.
“We simulate and model very large structures, systems and processes,” says Tajkhorshid, who also directs the National Institutes of Health Biomedical Technology Research Resource for Macromolecular Modeling and Bioinformatics. It’s a new type of computational biology that can be done on only the world’s fastest supercomputers. And Frontier will be mind-bogglingly fast –more than 1.5 quintillion calculations per second.
Like DOE, NIH has supported NAMD development for decades. More than 150,000 scientists worldwide use it and its sister program VMD (visual molecular dynamics). Tajkhorshid relies on NAMD to model fusion between a virus and its host cell, a major step in all viral infections.
“To ask questions about the (virus-host fusion) process” – which involves hundreds of millions or even billions of atoms – “we need to simulate it first,” he says. “Then we can look at the contribution of different components. What happens if you change the lipid composition? What happens if you don’t have a particular protein? What happens if you don’t have the right curvature of the membrane, and so on.”
Tajkhorshid’s team initially chose to study the Zika virus, spread primarily from mosquito bites, because there is no vaccine for it.
Frontier can provide the massive computational resources Tajkhorshid’s team needs to properly simulate the process. It involves calculating trillions of interactions of billions of particles for the biologically relevant span of at least a microsecond. “These are very demanding calculations,” he notes.
Tajkhorshid’s team initially chose to study the Zika virus, spread primarily from mosquito bites, because there is no vaccine for it. Pregnant women are particularly vulnerable; the virus can cause birth defects, including partial brain development.
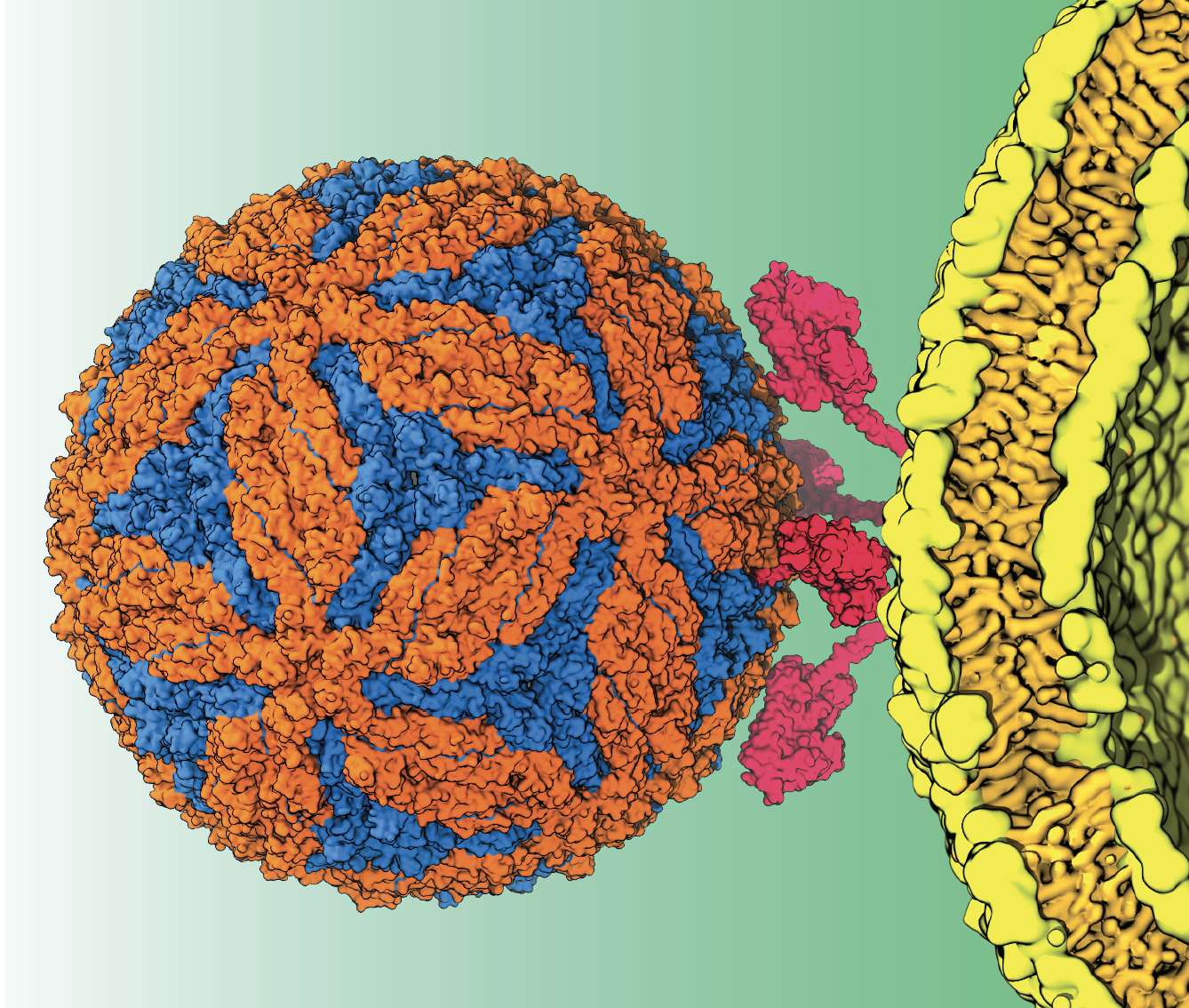
Zika also has been documented in detail. Already knowing the structure of the entire capsid – the protein shell enclosing the virus – and the virus particle itself has made it possible to begin studying how it enters host cells.
Additionally, Tajkhorshid says, “we have a membrane that sits on top of this viral capsid, formed when the virus comes out of the membrane of the host cell. So the virus inherits an extra membrane shell surrounding the capsid.” Some parts of the capsid penetrate the membrane partially or fully. The goal is to model both the membrane and the capsid together, and then to see how they enter the host cell.
Tajkhorshid has considerable experience in simulating membranes of complex structures and protein interactions. The Zika project posed a new challenge, however: to model the known shape of the entire capsid, then cover it in a membrane properly at the atomistic level.
Another challenge is deciphering how to fuse the virus membrane and the host cell membrane in a computer simulation.
“It is not trivial to bring two giant molecular pieces together in these molecular machines,” Tajkhorshid says. “We needed to implement several tricks in our modeling and simulation to be able to study this process. You have to fuse the two membranes to allow the genetic contents of the viral particle to go inside the host cell. We also have a very similar process in neurotransmission and hormone releases in the human body and in other organisms.”
Despite their incomplete understanding of the sequence, scientists do know that certain types of lipids and proteins play key roles.
“You need to have those lipids to be able to complete the process,” he says. “There are also certain proteins involved and if they are not activated the fusion will halt. These players have to join in the correct order and accurately; otherwise, we cannot do this.”
COVID-19 studies could possibly benefit from Tajkhorshid’s approach. His group already has nearly completed constructing a full model of the viral envelope that echoes coronavirus spike proteins.
“We might look at the coronavirus envelope binding to the host membranes,” he says. “The machinery, the approach, all the tricks that we need to do this for a coronavirus are going to be the same. In fact, for the coronavirus, it’s a bit less challenging because the viral envelope is less coupled to the proteins inside the virus. It’s a more isolated kind of object to look at.”
Tajkhorshid and many other scientists, meanwhile, eagerly await Frontier’s deployment.
“We’re greedy to simulate more complex and larger biological systems, a dream that can only be realized by having more capable machines.”