The DNA in every human cell gets damaged tens of thousands of times every day. A nucleotide – one of four basic units that comprise a chromosome – can get put in the wrong place while DNA is copied. Ultraviolet light from the sun can break DNA in two, and all kinds of chemical reactions can change the genes. DNA damage can stop the production of specific proteins or change them. Some of the damage can even be deadly.
At Georgia State University in Atlanta, chemist Ivaylo Ivanov creates computational models of various DNA processes – including repair.
“It’s extremely important that the genetic information is transmitted faithfully at each cell division,” Ivanov explains. That’s a crucial reason why processes exist to repair damaged DNA. Although Ivanov calls DNA “a relatively stable molecule chemically,” various compounds, even those produced in natural physiological pathways, can change the structure of a nucleotide. “DNA is subject to all these different chemical reactions, and all of these collectively can impact the transmittal of genetic information.”
Despite frequent damage to their DNA, most cells stay on healthy tracks and keep running normally. They “rely on elaborate repair pathways, biochemical pathways that fix damaged DNA,” Ivanov says. He’s interested in how that works, especially how it gets started.
With support from the Department of Energy (DOE) INCITE (Innovative and Novel Computational Impact on Theory and Experiment) program last year, Ivanov explored a key step in what the late Nobel laureate Francis Crick named the central dogma: the transcription process that uses DNA to make RNA and then proteins. The National Institutes of Health and the National Science Foundation also support his work. In particular, Ivanov looked at the beginning of making RNA from DNA.
DNA is transcribed into RNA and later serves as a template to build proteins. Enzymes called RNA polymerases control the DNA-to-RNA process. “Basically,” Ivanov says, “we were interested in figuring out how out RNA polymerases initiate transcription.”
First he gathered as much structural information as he could about the procedure. Most of that came from cryogenic electron microscopy, in which scientists image structures frozen at bitterly frigid temperatures, like that of liquid nitrogen, nearly minus 200 degrees Celsius. This technique rivals X-ray crystallography in revealing structural information.
Modeling the dynamics of transcription complexes requires simulating the movements of up to 1.5 million individual atoms.
Using the Summit supercomputer at the DOE’s Oak Ridge Leadership Computing Facility, open to all qualified researchers, Ivanov conducted molecular dynamics simulations based on these structural data. This, he explains, lets computational modeling simulate the dynamics of the complexes that participate in transcription. It only works, though, if there’s enough of the needed structural information from experiments. “The motions of the complex can suggest possible mechanisms, so you can make inferences.”
Scientists know from experiments that RNA polymerase plus other proteins assemble into a complex on DNA’s double helix. The RNA polymerase unwinds part of the DNA and separates the two strands, making what is known as a transcription bubble. Inside it, one strand of the DNA gets copied into RNA.
Modeling the dynamics of transcription complexes requires simulating the movements of up to 1.5 million individual atoms. The molecular dynamics method he uses tracks positions and momentum of each and every particle in a system, Ivanov says. “We integrate the Newtonian equations of motion and propagate the system to basically figure out the dynamics of the complex.”
The computational modeling makes up for some shortcomings of experiments, which capture only a few discrete states of the transcriptional assembly. With modeling, Ivanov tries to bridge those states to envision the process from start to finish – at least to the limits of what is being simulated. “But to what extent we would be able to accomplish this remains to be seen. We haven’t finished the calculation.”
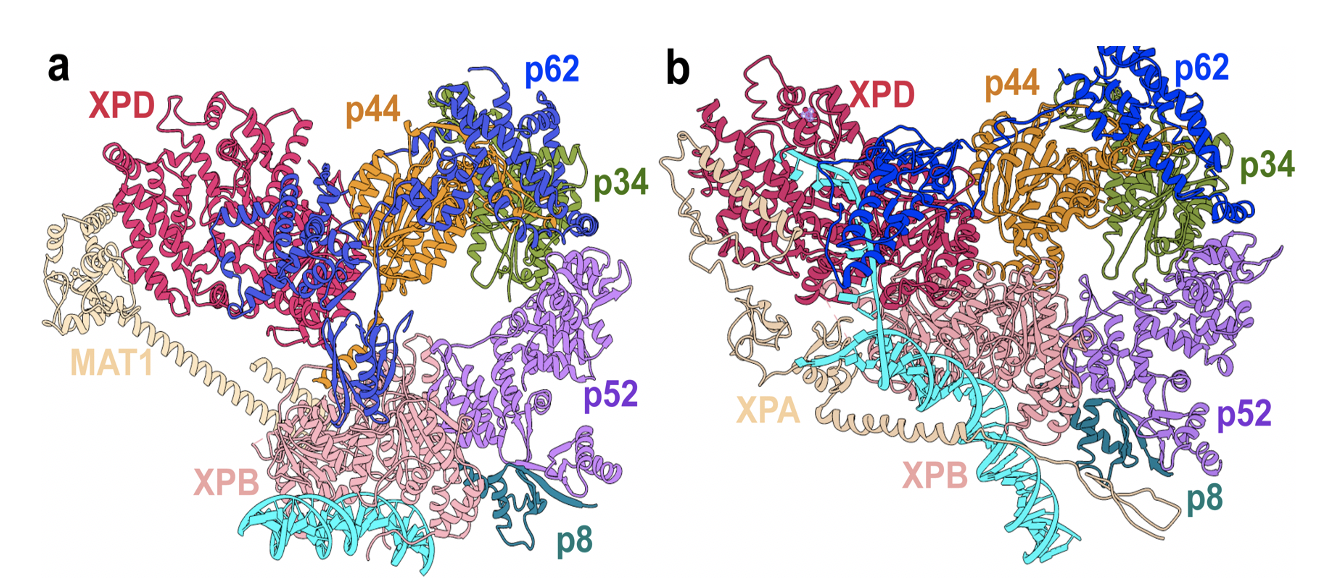
In that work, Ivanov looked in detail at transcription factor II H (TFIIH), which also plays a role in DNA repair.
With his INCITE award renewed this year, Ivanov will use Summit to explore TFIIH’s role in nucleotide excision repair. In this version of DNA repair, a damaged stretch of a chromosome is removed, and a repaired piece is put back in place. TFIIH helps initiate DNA translation to RNA and separately helps with nucleotide excision repair.
TFIIH consists of seven core components that form a horseshoe-like shape, with subunits called XPB and XPD at the ends. “The XPD and XPB are basically the components that are working on the DNA,” Ivanov notes. Other subunits serve as a spacer between the two, basically determining how far the horseshoe is open. There’s one subunit between XPD and XPB for transcription and another for nucleotide excision repair, and that subunit determines the shape of TFIIH, which in turn controls which job it’s ready to do.
As Ivanov describes it, “the nucleotide excision repair pathway is very interestingly coupled to the transcription process.” Even though TFIIH is responsible for both procedures, he says, “it shifts in shape when it’s participating in transcription initiation.”
Much like tracking the dynamics of TFIIH during transcription, Ivanov will apply similar computational modeling to this enzyme during nucleotide excision repair. One question that intrigues him: “What sort of communication do we have between the XPB and the XPD components?” He assumes that these components “act together to scan the DNA for damage and then unwind the DNA.” So he wants to model how that happens with dynamic simulations. “It’s kind of puzzling how one and the same component can perform these very different functions. It completely rearranges itself.”
Although lots of work lies ahead as Ivanov unravels just how TFIIH performs these different tasks, the results can help him understand what it looks like when these basic biology processes go right or wrong.
Missteps during transcription play a role in many diseases, from autoimmunity and cancer to cardiovascular disease and diabetes. Similarly, failing to repair damage in DNA sets off a range of cancers and inherited genetic disorders.
As Ivanov says, “all of this is tied to diseases.”