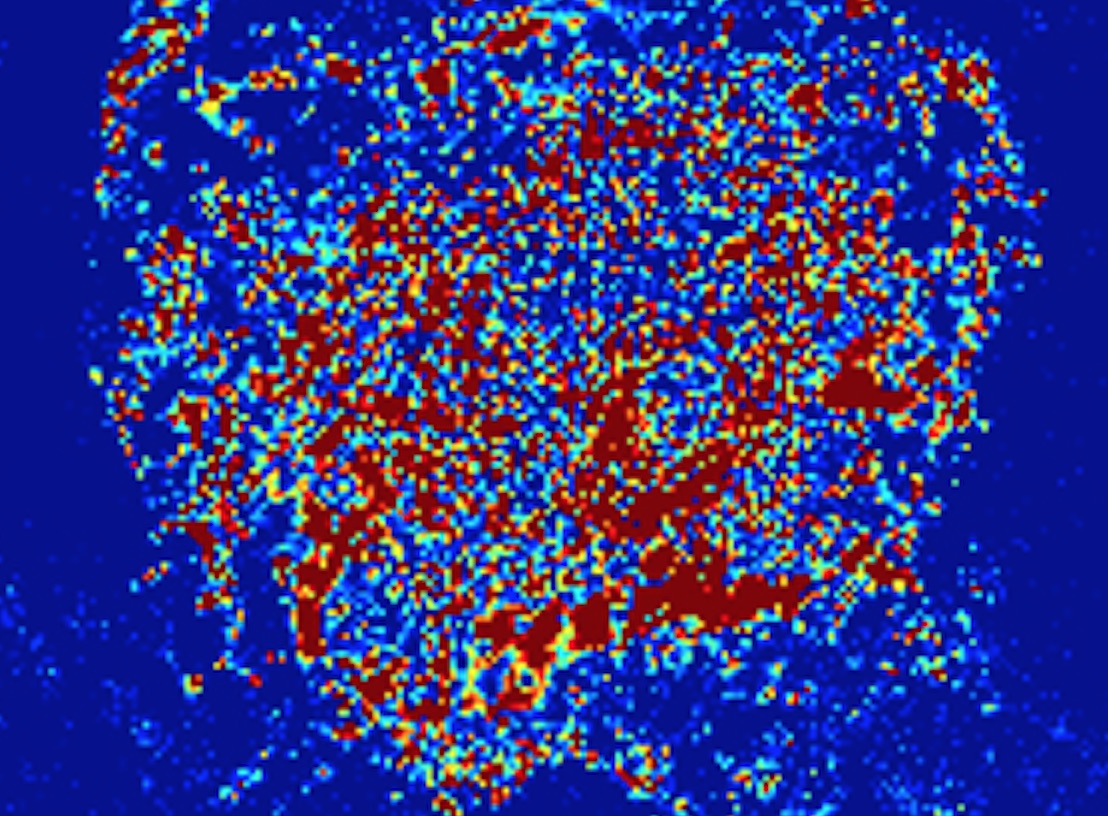
Northwestern University researchers reported a strange phenomenon earlier this year. The team, led by Vadim Backman, Walter Dill Scott Professor of Biomedical Engineering, had bombarded cancer cells in a petri dish with deadly ultraviolet (UV) radiation. Then they watched the cells die, using dynamic PWS (partial-wave spectroscopic) microscopy, a new technique that reveals the structure and motion of big molecule clusters, or macromolecules, inside cells.
In a thriving cell, macromolecules are always moving, huddling wherever needed to play their specialized roles in life-sustaining processes such as assembling new proteins or repairing damaged DNA. Dynamic PWS showed that, under UV light, this cellular machinery grinds to a halt.
At first, the macromolecules drift apart, forming a homogeneous intracellular broth. But after 10 to 20 minutes of UV exposure – and to the researchers’ surprise – a cell’s full complement of macromolecules bursts into a brief spasm of frenetic motion before the cell dies. The team named the phenomenon a cellular paroxysm. They think it points to a yet-to-be-found form of cell death and intend to explore how it might lead to future cancer treatments.
Only dynamic PWS microscopy could have made this discovery. The method detects intracellular structures of from 20 to 200 nanometers in size, too small to be seen with a conventional optical microscope. It does so without using labels or dyes, which can kill cells or disrupt their behavior. It follows whole molecule clusters, including elusive lipids and carbohydrates, not just specifically labeled proteins. And it records millisecond-by-millisecond motion within living cells in three dimensions.
Allen Taflove, a Northwestern professor of electrical and computer engineering, and his colleagues are helping develop dynamic PWS through simulations on Mira, an IBM Blue Gene/Q high-performance computer at the Argonne Leadership Computing Facility (ALCF), a Department of Energy (DOE) user facility at Argonne National Laboratory (ANL). The DOE Office of Science INCITE program recently granted Taflove a renewal award of 5 million node-hours on Mira to confirm further developments in the technology.
“Using the ALCF Mira resource, we validated the dynamic PWS microscopy technique and successfully applied it to study the time-domain response of cancer cells subjected to a lethal stimulus,” Taflove says.
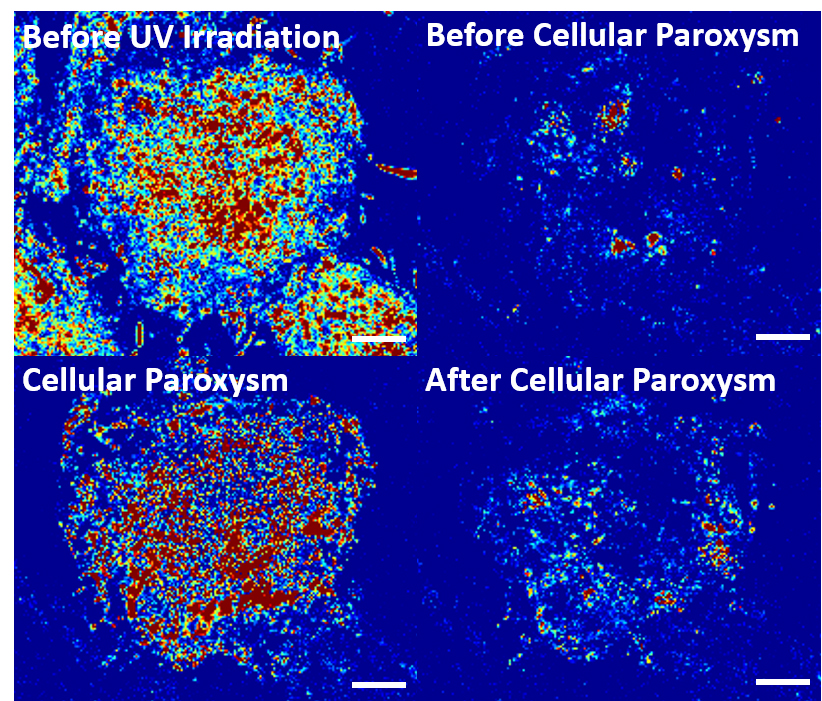
Cellular paroxysm (lower left) — a spasmodic reaction of molecules inside the cell —after several minutes of ultraviolet-radiation exposure before the cell dies (lower right). Work on the Department of Energy’s Mira supercomputer at the Argonne Leadership Computing Facility is helping fine-tune the microscopy technique as a new tool in cancer research. Image courtesy of McCormick School of Engineering, Northwestern University.
Taflove has collaborated with PWS creator Backman in his studies of early-stage cancer detection since 2004. Backman is a co-investigator on the INCITE award, along with Northwestern doctoral student Aya Eid and ANL computational science principal project specialist Wei Jiang.
Taflove’s contribution to Backman’s cancer research involves applying Maxwell’s equations to model PWS microscopy. Maxwell’s equations govern electromagnetic wave interactions with matter, including how visible light used in PWS interacts with the contents of biological cells. An electrical engineer, Taflove has pioneered finite-difference time-domain (FDTD) computational solutions of Maxwell’s equations since the early 1970s; his book on the topic is the seventh most-cited in all of physics. Beginning in 2007, he supervised the Backman lab’s decade-long development of Angora, an open-source, FDTD-based software suite that simulates both dynamic PWS and its predecessor, static PWS. It’s the primary code Taflove uses for his Mira INCITE awards.
PWS microscopy relies on three properties of light: backscattering, interference and spectra. A cluster of proteins or an unwound DNA strand isn’t large enough to produce an image in an optical microscope, but these macromolecules still scatter small amounts of light. Some of it bounces back, or backscatters. Light also backscatters off the petri dish’s glass surface. The macromolecules’ backscattered light interferes with that from the glass surface, resulting in areas of altered spectra. As observed in the image plane, PWS measures the spectrum of each pixel arising from this interference. Analyzing the pixels’ spectra lets researchers characterize the macromolecular structures’ statistical nature. PWS doesn’t render actual images of individual macromolecules. Instead, it provides information about whether the structures are distributed homogeneously or in clusters.
Dynamic PWS builds on the distinguished history of static PWS. After Backman’s team invented the technique, Taflove and colleagues used detailed Angora computational modeling to help validate it in 2013. Then Backman’s group used static PWS to study macromolecular structures in more than 800 cancer patients and control subjects. The group showed that a phenomenon called the field effect, or field carcinogenesis, can be used to detect cancers much earlier than current methods and without a biopsy. No one fully understands the field effect, but scientists have shown in recent decades that cancer cells affect normal-appearing cells around them.
Defying skeptics, Backman and his team detected malignant tissues by finding abnormalities in nearby normal-appearing tissues. A more random macromolecular structure indicated a higher cancer risk, even in cells collected some distance from a tumor. For example, lung cancer’s signature appears in cheek swabs and ovarian cancer’s mark shows in Pap smears. Over the past decade, the team has quantified cancer risk in these and five other organ types: colon, pancreas, esophagus, prostate and thyroid.
‘The simulations on Mira were much closer (than analytical theory) and showed the correct trend.’
Now Backman’s team has created dynamic PWS – adding the element of millisecond time resolution to their tests. They intend to search for more early signs of cancer based on abnormal movements of cellular macromolecular complexes.
To validate dynamic PWS, the team ran experiments in parallel with Angora simulations on Mira. The experiments used uniform-size nanospheres in place of cellular macromolecules. Like the much-larger phantom objects used to calibrate MRI and CT imagers, the nanosphere phantoms’ properties also are precisely known. In place of a glass petri dish, the team used another material to make the mismatch of refractive properties comparable to the mismatch between glass and cellular macromolecules.
Finally, the team compared the experimental results with two predictions, one from the simulation and one from physics-based analytical theory, Taflove says. “The analytical theory was initially not quite in accord with the experimental data obtained from the nanosphere phantoms, whereas the Angora simulations on Mira were much closer and showed the correct trend. The Angora simulations performed on Mira for the nanosphere phantoms ultimately did get the analytical theory rounded into shape.”
Once the team refined its theory, it was ready to demonstrate the technique in three studies. In one, the researchers confirmed that dynamic PWS can capture information about both motion and structure in living cells without using chemical labels. They applied paraformaldehyde to cancer cells in vitro. In this widely used laboratory technique for fixing cells, chemical crosslinking occurs among large molecules, halting cellular activity. Using dynamic PWS, the team observed the cells as they underwent crosslinking cellular fixation. As expected, the macromolecular structure remained heterogeneous, or diverse in composition, as crosslinking bonds stopped the motion and locked the structures in place.
The second study also yielded expected results. The team showed that genetic material in undifferentiated stem cells is more heterogeneous and mobile than in differentiated cells. More active nuclei make sense for stem cells, which need access to a wide range of genes as they develop into various cell types.
In the third study, the researchers tracked changes in macromolecular motion that occur during UV-induced cell death. They described the process of cell death in detail and discovered cellular paroxysm.
The Backman group’s next steps will involve dynamic PWS studies of how potential novel therapies produce transient responses in cancer cells. Can the newly observed phenomenon of cellular paroxysm be induced medically for each and every cancer cell in a tumor? Dynamic PWS will help reveal the answer.
“Solidly based upon the fundamental physics foundation of Maxwell’s equations, millisecond-resolution dynamic PWS is a proven tool for tracking macromolecular motion within living cells,” Taflove says. “Now, let’s apply it to investigate novel potential cancer therapies.”