When biologists needed to quickly analyze strains suspected in the E. coli outbreak still reverberating through Europe, the Department of Energy’s Argonne National Laboratory tapped into its Magellan cloud computing testbed.
Genetic analysis that usually would have taken weeks was compressed to one weekend in June, allowing scientists from around the globe to rapidly scrutinize the food-borne bacteria believed to have killed dozens and sickened thousands.
Magellan, Argonne’s Rick Stevens says, “enabled us to dynamically scale up the capacity to keep ahead of the demand that was coming from all over the world.”
The machines that comprise Magellan(see sidebar, “The nuts and bolts of Argonne’s cloud”) give researchers an arena for testing cloud computing’s utility for scientific discovery. Projects too small for DOE’s supercomputers but too large for in-house desktop or cluster computers can run through Magellan as needed. By creating a virtual machine, researchers can choose the number of processors for their applications.
Magellan and Argonne were part of an international network that sprang up to trace the deadly bacterial strains’ genealogies and to seek their weaknesses. The Argonne group quickly grasped Magellan’s potential, says Stevens, Argonne’s associate director for computing, environment and life sciences and a senior fellow in the lab’s Computation Institute.
Driven by social networking tools, “the labs in Europe had much more of a sense of urgency” than American labs, Stevens says. “We were trying to find some unique way to help.”
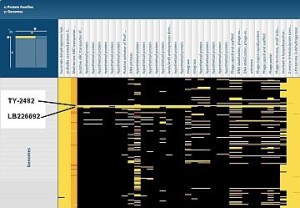
Virginia Bioinformatics Institute scientists compared proteins from two new pathogenic E. coli strains, LB226692 and TY-2482, with proteins from other bacterial genomes using the Protein Family Sorter on the Pathosystems Resource Integration Center (PATRIC) website. This shows part of a map, with several unique islands of proteins identified in the two genomes. Click the image to see the complete map. Credit: PATRIC website.
Stevens belongs to a group that developed the RAST (Rapid Annotation using Subsystems Technology) server, a longstanding program that can sift through a bacterial genome and recognize features in DNA sequences, such as where genes start and end or regions that code for proteins or regulate genes. The system “uses a combination of methods to make a best guess or best estimate of what the function is,” Stevens says. It compares DNA segments to a database of known gene functions to find similarities (such as chromosome locations of other organisms) and candidate proteins to known proteins or protein families.
Argonne hosts the free RAST server with support from the National Institute of Allergy and Infectious Diseases (NIAID) Pathosystems Resource Integration Center (PATRIC). Bacterial genomics researchers from all over the world log on and submit sequence data, then retrieve an annotated genome 12 to 24 hours later. RAST has annotated around 23,000 genomes over the past two years.
Two new genomes came in on Thursday, June 3. Scientists in China and the United States, working with German researchers, had sequenced the DNA for E. coli strains designated TY-2482 and LB226692. Both were suspects in an outbreak of food-borne illness that surfaced in late May.
The ailment spread quickly, mostly in Germany, and was noteworthy for its virulence: An especially high percentage of victims developed a condition that can cause kidney damage or death. Within a month, the infection – ultimately traced to contaminated raw bean and seed sprouts – had sickened more than 3,800 people and killed 48, the World Health Organization reported.
Stevens and his ANL colleagues saw a sudden spike in activity on the RAST server late on June 3, soon after the first DNA sequence, for TY-2482, came out. Researchers were submitting E. coli genomes for annotation and comparison with the outbreak E. coli. Using Twitter, Facebook and other tools, scientists from around the world connected with each other, spreading the news that the sequence data were available, Stevens says, and research groups began spontaneously analyzing it.
Meanwhile, PATRIC scientists at Virginia Tech’s Virginia Bioinformatics Institute (VBI) integrated the new RAST-annotated outbreak strains with the publicly available annotated E. coli genomes in their website’s database for comparison.
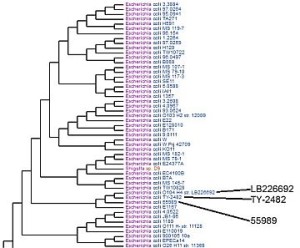
Virginia Bioinformatics Institute scientists compared the genomes of E. coli strains LB226692 and TY-2482 with other bacterial genomes in the database of the Pathosystems Resource Integration Center (PATRIC) to develop phylogenetic trees. By looking at the genes in each strain and what proteins they code for, researchers can tell how closely they are related. This tree shows the two strains, suspected in an outbreak of food-borne illnesses in Europe, are most closely related to strain 55989, a pathogenic type that adheres to the walls of intestines. Credit: PATRIC website.
“Within an hour or two, we had 200 or 300 genomes submitted,” Stevens says. Since each annotation and comparison takes hours, the Argonne researchers quickly recognized that RAST would bog down. They turned to Magellan.
Magellan is designed to investigate scientific research using cloud computing, which makes a shared pool of computer resources available. Users access processors, software and other assets via networks, allowing them to quickly increase or cut capability as needs change. Although clouds are increasingly common in industry, most researchers are only beginning to use them.
The RAST team had already tried using Magellan to create virtual versions of the server to increase throughput. Now the sudden, urgent demand for annotated genomes was an opportunity to scale up those tests.
“The idea was to have enough capacity so turnaround time for the E. coli jobs could be as small as possible – so the whole community could get a rapid response,” Stevens says. The Argonne team created multiple virtual versions of the server on Magellan, each with an operating system, applications and data to do annotation and analysis. “Think of it like this: Within a few minutes we multiplied the server 60 times to give it more capability. That kind of on-demand scaling up is exactly what clouds are for.”
As updated versions of the German E. coli genomes became available, biologists annotated and analyzed them, quickly gobbling up the added computing power. “The number of jobs kept ramping up,” Stevens says. “The more people became aware of it, the more they had ideas and were submitting things.”
The Argonne and Virginia Tech teams worked around the clock the weekend of June 5 and 6, monitoring activity and doing their own analyses. PATRIC saw a 50 percent jump in activity, most of it from researchers downloading annotations of the German E. coli and using the site’s analytical tools, says VBI’s Ron Kenyon, who manages the project.
VBI works with Argonne to compile RAST annotations in PATRIC, a clearinghouse that compiles biomolecular data – including genomes – for pathogenic bacteria. “We use RAST for annotating all the genomes so you have a consistent, apples to apples comparison,” Kenyon says.
The VBI researchers did their own analyses to trace the deadly bacteria’s heritage. E. coli strains, most of which are benign, often swap chunks of DNA, creating new strains capable of generating various proteins, some of which cause illness. Most of the German strains’ interesting features were associated with mobile elements, Stevens says, including factors for antibiotic resistance, for producing the toxin that sickens people and for making the bacteria adhere to things – like vegetables.
“One of the challenges was trying to understand where those genes come from. They’re not in all E. coli strains,” Stevens says. Over the weekend of June 4 and 5, VBI researchers compared the German strains’ annotations with annotations for about 200 publicly available E. coli genomes. Phylogenetic trees show how closely different strains are related – “where they fit from an evolutionary standpoint,” Kenyon says.
The phylogenetic trees showed the two strains are closely related to a pathogenic enteroaggregative strain, Kenyon says – a disease-causing E. coli that blankets the intestinal lining and leads to severe illness and death. VBI researchers are continuing to release results from studies of the German strains.
Throughout the weekend, researchers from around the world continued probing genetic data for the German strains. “There was a whole range of computation done, after the initial burst, that was trying to fill in the details,” Stevens says. “There was a lot of excitement.” Scientists “kept thinking, ‘What else can I do? I’ve got these resources here. Maybe I’ll run this analysis. Nobody’s tried this yet.’” It was perhaps the first time a global research community had spontaneously coordinated activities via social networks.
With their results, researchers can find unique aspects of the deadly strains and create tests to identify them. They can find which antibiotics the bacteria resist and which will kill them. And doctors can use information on toxins the bacteria produce to devise treatments.
“You’ve got the blueprint of the enemy,” Stevens says. “Now you’ve got to figure out how to attack it.”
