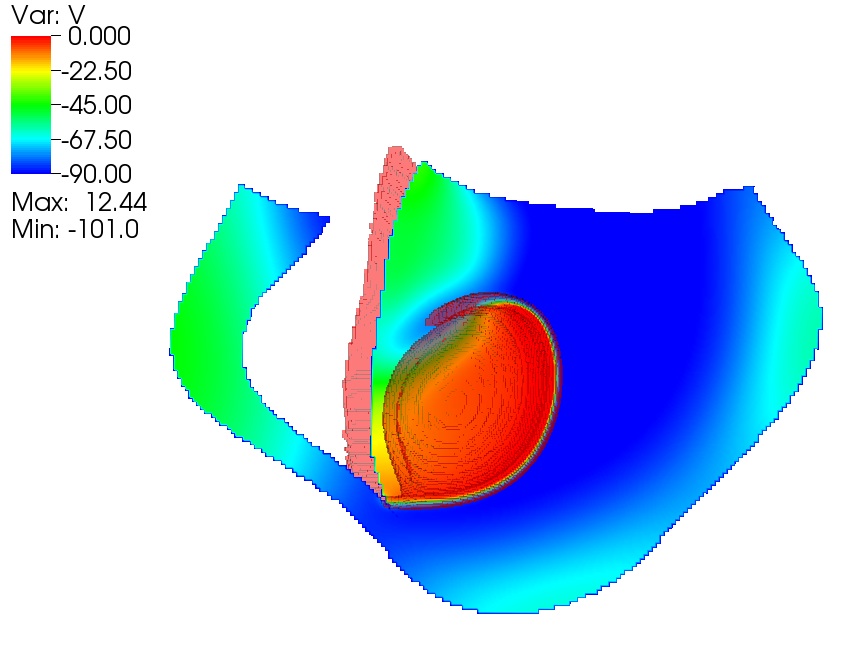
The rhythm of a human heart, beating along at 70 or so contractions a minute, arises from precisely timed electrical waves. These waves must travel the entire three-dimensional shape of the heart to activate the muscles in a sequence that pumps blood throughout the body.
Unfortunately, human hearts do not always beat just right. In fact, millions of Americans suffer from irregular heartbeats, or recurrent arrhythmias. The most serious arrhythmias can cause sudden cardiac death.
The intricacies of these waves and their link to health problems caught the attention of Jeffrey Fox, a visiting scientist at Cornell University’s Center for Applied Mathematics. As a Cornell graduate student, Fox had worked with physics professor Eberhard Bodenschatz on nonlinear dynamics and pattern formation and with veterinary medicine professor Robert Gilmour, who studies the physiology of cardiac-rhythm disorders. This combination led Fox to research chaotic dynamics in cardiac tissue.
When operating normally, the heart’s sinus node makes a pacemaker electrical signal that generates a smooth, plane wave that travels through the tissue. If that wave hits diseased or dead heart tissue, though, the plane wave can break into spiral waves.
“The rapidly moving spiral waves are like a hurricane in the heart,” Fox says. The spiral waves break into many waves and regions of the heart contract out of sync. “So the heart wiggles instead of pumping.”
Fox and his colleagues are using combinations of mathematical modeling and computer simulations to study the cause of such spiral waves and the resulting cardiac arrhythmias.
At Cornell, Fox met Colin Hill, who was working on a graduate degree in physics. They started collaborating to model genes. That research led Fox and Hill to the university’s supercomputer, which had 300 processors at the time, making it one of the fastest machines outside the military.
The computer’s speed and power, at least for its day, drew heavy use, Hill says. “We had to stay up till 2 a.m. to run on most of the processors.”
The work at Argonne is ‘game-changing science,’ Hill says.
The gene-modeling work was just the beginning of their collaboration. In 2000, Hill and Iya Khalil founded Gene Network Sciences (GNS) in Cornell’s home town of Ithaca, N.Y. There Fox continued his research, focusing on canine heart simulations.
Fox’s heart simulation work is based on a range of modeling strategies. For example, he uses simple mathematical models to pull out the key determinants of how electrical waves move through the heart. In addition, he models ion channels, the pores that control the electrical charge across a cell membrane. Currents flowing through these channels can be calculated with the Hodgkin-Huxley equations, which earned Alan Lloyd Hodgkin and Andrew Huxley the 1963 Nobel Prize in Physiology or Medicine.
These ion currents, though, make up just part of a cardiac-cell model. It includes ion channels that are voltage gated – opening or closing based on the charge across the cell membrane – and ones that are not, as well as ion pumps and more. All of that information gets expressed as a differential equation that summarizes about a dozen variables and about 60 parameters.
Then lots of these cardiac cells get combined in a 3-D model of heart tissue. “Partial differential equations are used to model the cell-membrane potential changing in space and time – the wave moving,” Fox says. “To solve that numerically, we have to take a big piece of the heart tissue in space and cut it into little pieces.” But with luck, putting all of that together will build the emergent phenomenon of a heartbeat.
“It’s complicated in space and time,” Fox says. To make it a little simpler, Fox models only the canine-heart ventricles – the two lower chambers that pump blood out to the lungs and body.
Hill’s work depends on multiscale modeling, which portrays phenomena over large spans of space and time. The GNS platform does just that.
“We didn’t add multiple dimensions (to our platform) as an afterthought,” Hill says. “We designed our platform to be very much agnostic to actual models.” That flexibility makes it possible to plug the heart model into the platform.
But that doesn’t mean it’s easy. “Fitting the parameters has been the biggest challenge,” Hill says.
The heart simulation also faced another challenge: a demand for computational speed. Luckily for Fox and Hill, today’s hardware is much more powerful than the 300-processor supercomputer they first used. “Now we routinely plug into machines with tens of thousands of processors,” Hill says.
In 2008, Fox and GNS scientists got a chance to plug into a really super supercomputer – Argonne National Laboratory’s IBM Blue Gene/P. Through its INCITE (Innovative and Novel Computational Impact on Theory and Experiment) program, the Department of Energy’s Office of Science granted GNS an award of almost 850,000 processor hours on the computer, dubbed Intrepid. In 2009 their INCITE grant was renewed and increased – to almost 21.5 million processor hours.
One benefit of working on the Blue Gene/P is its scalable architecture. “We can test code on a system running 2,000 processors, and then port that to a 40,000-processor machine,” Hill says. “We don’t need to do more testing to make the machine work.”
To make Fox’s cardiac simulation run fast, his computational colleagues assign different regions of the ventricles to different computer processors. “Each processor is doing equations for its part of the heart,” Fox says, “and then passing information to processors doing neighboring regions of tissue. Blue Gene is hardware designed to do that, and very effectively.”
The GNS team typically used two racks, or nearly 8,200 processors, when running heart simulations on the Blue Gene/P. The largest run used eight racks, verging on 33,000 processors. Using more racks made the simulations faster, but also meant that the researchers waited longer in the queue until enough processors were available. So Fox and his colleagues would use fewer racks for easier simulations but wait for more racks when they needed the additional computing power.
On Blue Gene/P, Fox and his colleagues modeled the activity of two ventricles of a canine heart for 10 seconds or so in chunks of one or two seconds. This provided roughly 25 cycles of activation – each taking 400 to 500 milliseconds during typical beats but occurring much faster during arrhythmias. Modeling a single activation only took about 10 minutes on Intrepid, but it would have taken two months or so of solid runtime on the GNS 32-processor cluster. “We couldn’t have done this work without the Blue Gene/P,” Fox says.
The GNS team achieved more than simply modeling those waves. They also challenged their model to replicate a real-world result. Cornell’s Gilmour knew of a specific set of conditions that creates ventricular arrhythmia in a dog. When Fox and his colleagues created the same scenario in their simulation, it generated spiral waves.
The work at Argonne is “game-changing science,” Hill says. “We can simulate the canine heart in a way that was not possible before” and can study new cardiac frontiers.
For example, their model could be used to predict how the heart would respond to a particular drug. Fox asks, “If we change the properties of ion currents – like a drug that binds one of the channels – how does that change wave propagation?”
In the April 2009 Annals of Biomedical Engineering Fox and several GNS scientists describe using some of their modeling to test the effect of a drug on the so-called rapid delayed rectifier potassium current, or I(Kr), which is commonly used as one of the cardiac-safety indicators.
In that paper, the authors conclude that the method has the potential to produce data-driven models that reproduce ion channel behavior in heart cells. Say the authors, “Although the method was developed for I(Kr), the same strategy can be applied to other ion channels… .”
The GNS heart simulation also could lead to improved medical devices. Cardiologists already implant devices – pacemakers – in arrhythmia sufferers, but this simulation could help to enhance future products.
“Can we make smarter devices that might detect trouble before this very complicated electrical storm has started?” Fox asks. “If we can, then maybe we can use a small shock that is a little smarter.”