Just call him the “Glue Guy.”
No, Chris Oehmen doesn’t have an Elmer’s fixation. He’s the glue that helps hold together research collaborations spanning disciplines from microbiology to chemistry to high-performance computing.
Oehmen’s earned his glue-guy status by articulating the  needs of each scientific branch. He can approach a research problem from a computational standpoint and explain its requirements to a biologist, then turn to a computer programmer and explain why the biologist needs to see data output organized in a specific array.
needs of each scientific branch. He can approach a research problem from a computational standpoint and explain its requirements to a biologist, then turn to a computer programmer and explain why the biologist needs to see data output organized in a specific array.
Oehmen bridges both worlds with an ability to speak the languages of biology and computing. It’s a skill he gained during a Department of Energy Computational Science Graduate Fellowship.
Since joining Pacific Northwest National Laboratory in 2003, Oehmen has lent his talents to one of the grand challenges of 21st century biology: making sense of mountains of data relating to genes and the proteins they encode, then using that data to generate biologically relevant discoveries.
Oehmen led the Pacific Northwest lab team that earned a top-three place in the Supercomputing 2006 Analytics Challenge, a worldwide contest showcasing the best examples of using high-performance computing to solve real-world problems. The team performed its work at the Molecular Science Computing Facility at the Environmental Molecular Science Laboratory, a DOE national scientific user center.
For the project, Oehmen collaborated with Bobbie-Jo Webb-Robertson, a Pacific Northwest lab senior research scientist, microbiologists Lee Ann McCue and Joshua Adkins, and with programmers, data analysts, mathematicians, and graphics experts.
They analyzed biochemical pathways in the bacteria Salmonella, an organism that causes food poisoning. The researchers zeroed in on “pathogenicity islands” — DNA segments that produce many proteins responsible for making a microbe toxic, or “virulent.”
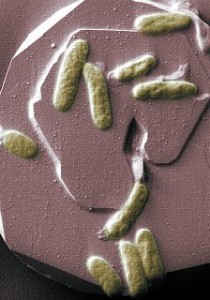
PNNL scientists are applying systems biology methods to understand electron transfer at the microbe-mineral interface in the model organism Shewanella oneidensis MR-1 (shown above). Such microorganisms exchange electrons with minerals in soil, sediment, and subsurface material, and could biologically remediate uranium contamination.
Using statistics and visualization tools Webb-Robertson and others developed, the team identified proteins present in the virulent growth condition that were not present in the non-virulent growth condition, validating the group’s analytical toolbox.
“Not everyone has access to high-performance computing,” Oehmen says. “What we were trying to do is bridge the gap so that through our visual analytics component, scientists could have access to the computational power of a supercomputer from their desktop.”
Biologists require computational power for completely different purposes than physicists and engineers, Oehmen says. Physicists often construct formulas based on well-understood physical principles and use them to describe large dynamic systems, such as fluid flow or heat conduction.
Biologists, in contrast, deal with many layers and levels of data simultaneously, he says. Their problems are analogous to data-mining problems.
To answer that kind of question, Oehmen has joined scientists from the Pacific lab and Oak Ridge National Laboratory in a project dubbed DICCBS — Data-Intensive Computing for Complex Biological Systems.
The group, led by Tjerk Straatsma at Pacific Northwest and Nagiza Samatova at Oak Ridge, is creating data-analysis, modeling and data-mining tools to extract knowledge across layers of biological information, from genes to proteins to whole organisms.
As part of the project, Oehmen has adapted the analytic program BLAST, a nearly universal code biologists use to make gene sequence comparisons. Oehmen modified the code to run on a parallel computing platform, increasing the efficiency of searching genomic data by 1,500-fold.
“I can’t imagine working anywhere else,” Oehmen says. “We have a critical mass of people doing pure science and bringing new technology together to solve the biggest problems in biology. What could be better than that?”