Part of the Science at the Exascale series.
Simulating biology won’t be extremely useful without extreme-scale computing. Biology demands a whole-systems view to study life and living things deeply.
Whether “looking at the atomic structure of molecules or ecosystems, you run into a rather astounding level of detail across large expanses,” says Mark Ellisman, professor of neurosciences and bioengineering at the University of California, San Diego. “Biologists face the largest data challenge in modern science.”
The most challenging questions in biology require computing power “an order of magnitude beyond what we have today, and that means reaching into the exascale,” says Susan Gregurick, program manager for computational biology and bioinformatics at the Department of Energy’s Office of Biological and Environmental Research.
A big area in energy research, she notes, involves molecular-dynamic simulations that show how enzymes break down cellulosic feedstock into fermentable sugars for biofuel production.
The models we have now are simplistic and at a smaller scale than needed, she says. “We have to extrapolate based on modest-size simulations.”
As computing power grows, researchers can increase both the scale and the complexity of their simulations.
“We want to explore everything, such as understanding how pretreatments – like acid hydrolysis – affect cell-wall materials and their digestibility into biofuel components.”
It will take a big advance in computation to get where Gregurick wants the field to be. “There are no more incremental changes that will have a big impact.”
When the big impact arrives, it will be wide-ranging.
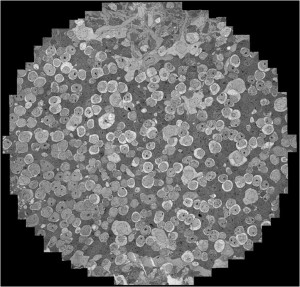
University of Utah researchers combined thousands of electron micrographs to create a 3-D map of the cells in a rabbit’s retina. Image courtesy of Chris Johnson.
With sufficient advances in computer power and algorithm development, for example, the generalized medicine doctors practice today could be tailored to specific patients and their unique biology, says Chris Johnson, director of the Scientific Computing and Imaging Institute and distinguished professor of computer science at the University of Utah in Salt Lake City.
Researchers could one day make functional computer models of individual anatomies, Johnson says. “A patient will get an MRI or CT scan, and then we can use those with image analysis to pull out and characterize different tissue types and even more detail, such as the boundaries between scalp and skull in the brain.
“We can use segmented images to make geometric models based on the patient’s anatomy, then do large-scale simulations to better understand function, such as electrical activity in the brain or heart. Maybe we can customize a hip implant or other implant.”
The computational horsepower needed, however, depends on the problem. The heart, for example, has about one billion cells, Johnson says.
‘We capture the essence but leave out some of the unnecessary complexity.’
“Each cell has about 30 ion channels, and we need to understand the flow of ions. That makes 30 billion degrees of freedom, and then you deal with that every millisecond. To get a few seconds, that turns very quickly into a large-scale problem.”
And that’s only looking at the electrical aspect, he notes. To simulate heart movement adds another problem of similar size.
“At every level, it’s challenging.”
In the push to run biological applications on extreme-scale computers, bottlenecks could emerge from many elements: hardware, software and algorithms – “throughout the stack,” says Anne Trefethen, director of the Oxford e-Research Centre.
For software, from operating systems through algorithms and applications, she says, “there’s lots of work to be done. The software that we have at the moment would not scale to what we need. We must look at the algorithms we are using to be sure that we could use the full breadth of an exascale computer.”
Johnson says that’s key to increasing the capability of computational biology.
“With every new set of hardware, we need software that can efficiently and effectively implement the algorithms on that hardware. It’s a lot harder than you would think. The software side is lagging behind our ability to create faster hardware.”
Computer power and algorithm development have progressed enough over the past 10 to 15 years that scientists can make useful biological and physiological simulations, Johnson says, but they’re still simplified.
“We capture the essence but leave out some of the unnecessary complexity. You can only simplify to a certain point, though, and still get useful and adequate answers.”
Computing power is needed to understand those answers – and the need will only become greater as more and more information pours forth from both simulations and experiments.
“We need visual data analysis,” Johnson says. Visualization tools must simplify data or abstract them. “We need to transfer those data into information that our brain can understand.”
Developing software for massively parallel supercomputers usually starts small, with “toy problems that you can get your brain around,” Johnson says. “We might find good ideas from these. Then scale that up to enormous problems.”
Scaling up to extreme computing, however, will trigger its own set of difficulties, Trefethen says. “With an exascale computer, there are many millions of processors, so we must deal with the fact that processors will die. The software needs to cope with that, adapt to it.”
Will biologists move into exascale computing with general tools or will they need unique ones for each application?
“As we go forward with looking at software for exascale – coding methodology, understanding the requirements of applications – we must see how we will adapt software and maybe hardware,” Trefethen says. “At the extreme, you could end up with software that is suitable for only one application.”
But because achieving exascale will require a large investment, researchers are looking at tools and infrastructure that will support classes of applications.
“Only a few applications will truly be able to scale to exascale at first. But if we get it right, we will have the infrastructure there to support more applications for future generations. We hope to be able to solve generic issues for a relatively broad range of applications.”
Johnson hopes for some crossover between problems. “A lot of science, engineering, and biomedicine have underlying physical relationships, so we can create some general techniques and visual data analysis that can apply across those fields.” Despite that, he says, “there is no one-size-fits-all solution.”