At the GE Energy Learning Center in upstate New York, workers learn to fix and maintain wind turbines. Physicist Masako Yamada works nearby, at the GE Global Research Center.
Whenever she passes the learning center, Yamada recalls a nacelle, a bus-sized hub she’s seen on the training center floor. Generators housed in nacelles atop wind-turbine towers convert into electricity the rotation of blades longer than semitrailer trucks.
It’s a reminder of the huge impact Yamada’s studies of tiny processes could have. At the research center, she focuses on the molecular foundation of the freezing process, helping find surfaces to keep turbine blades ice-free and efficient.
“Ice formation is a really complex problem,” Yamada says from her office in Niskayuna, N.Y. “If it were just a surface standing still, that would make things easier, but by the time you add impact, humidity, wind – all these things – it becomes much more difficult.”
The Department of Energy’s Advanced Scientific Computing Research (ASCR) program is helping, providing Yamada and her colleagues 40 million processor hours to tune and run their freezing simulations. The ASCR Leadership Computing Challenge grant gives them access to Oak Ridge National Laboratory’s Titan, a Cray XK7 rated one of the world’s top supercomputers.
Industry studies show that wind turbines can lose from 2 to 10 percent of their annual production to ice-related slowdowns and shutdowns or the cost of heating blades to shed ice, Yamada says.
For years, researchers have tested hydrophobic (water repellant) and icephobic materials – including ones incorporating nanoscale textures thousands of times smaller than the width of a human hair – but none is completely satisfactory or practical.
“Nanotechnology offers promise on the lab scale, but we need to consider manufacturing processes that can cover 50-meter-long surfaces,” Yamada says. And “the texture has to be durable for years under very harsh conditions.”
‘What we’re trying to do is map out this parameter space and learn what solutions work best for given sets of environmental conditions.’
A good ice-shedding surface for wind turbines must perform well in and withstand a variety of temperatures and humidities. It must survive long-term ultraviolet light exposure and blows from sand, insects, water droplets, snowflakes and ice crystals.
The surfaces Yamada and her colleagues develop could have a range of applications beyond wind turbines, from windshields to industrial structures like oil derricks.
To understand freezing, scientists first must unravel nucleation: the first hint of ice formation at the molecular level. “You have to have a tiny, tiny seedling to cause the ice to form,” Yamada says. Ice nuclei can form around a dust speck, as in snowflakes, or water molecules can spontaneously rearrange themselves to form a nucleus.
“If you can keep those seedlings from forming, you can keep ice from forming,” she says. Her computer models use physical principles to calculate in detail how different conditions affect nucleation and what slows or stops it. The data she and her colleagues produce help guide researchers to test the most promising ice-resistant materials and conditions.
Real-life wind turbines “can run at all sorts of temperatures and all sorts of humidities and all sorts of impacts,” she says. “What we’re trying to do is map out this parameter space and learn what solutions work best for given sets of environmental conditions.”
To answer those questions, Yamada uses molecular dynamics (MD) codes to track individual water molecule movements over time.
MD methods’ time dimension lets researchers ask “when does this nucleus form, how does it form, where does it form?” Yamada says. Experiments can’t probe every molecule’s position in time increments of femtoseconds – quadrillionths of a second. “That’s why molecular dynamics is used as a supplement to help us understand the things we can’t see.”
The difficulty: Each molecule interacts with every other one, making the problem substantially more demanding as molecules are added. Even cutting-edge models portraying submicroscopic droplets for spans far shorter than an eye-blink require millions of compute-time hours distributed across thousands of processors.
Titan can do the job. Its heterogeneous architecture combines nearly 300,000 standard central processing units (CPUs) with more than 18,000 graphics processing units (GPUs) to accelerate calculations. The approach gives Titan high speed for less electrical power, but complicates programming.
Yamada has adopted recent advances made by researchers across the country to increase the simulations’ efficiency. ORNL’s W. Michael Brown helped tweak LAMMPS, a popular MD code developed under the auspices of Sandia National Laboratories, so a key algorithm could take full advantage of Titan’s GPUs. As a result, a one-million-molecule water drop simulation ran five times faster than a standard version previously benchmarked on Titan’s predecessor, Jaguar.
Working with Arthur Voter at Los Alamos National Laboratory, Yamada also is implementing the Parallel Replica Method for these MD calculations. In essence, it runs multiple copies of an identical simulation to more quickly reach the desired information.
“Grossly speaking, if I ran 100 identical simulations I’d be able to see freezing in hundredth of the time. That’s the premise.” Sandia National Laboratories’ Paul Crozier also helped adapt LAMMPS to better distribute the workload of calculating the droplet’s diverse structure over the computer’s many processors.
Another improvement: The code uses a more efficient water model, developed by Valeria Molinero’s research group at the University of Utah.
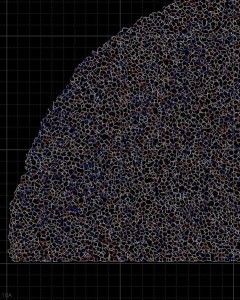
In this close-up of a visualization of a million-molecule water droplet simulation, lines indicate bonds among the water molecule models. White denotes average particle mobility, red denotes higher-than-average particle mobility and blue denotes lower-than-average particle mobility. Image courtesy of Masako Yamada, GE Global Research.
Taken together, the software alterations alone have increased the simulation’s efficiency by hundreds of times, Yamada says. “That’s what I’m really excited about, because we’re seeing these orders of magnitude productivity improvements” in the span of a few years.
Improved codes and access to Titan have opened new possibilities for Yamada’s research. As a graduate student in the 1990s, she used high-performance computers to run MD simulations of 512 water molecules. Now she typically models a million molecules.
The simulations also can run for more time steps, portraying molecular behavior for a longer period. That’s especially important for simulations of freezing.
“Molecular dynamics is wonderful when the temperatures are hot and the processes are fast, but when the temperatures are cold and processes are slow you have to wait a very long time to observe anything,” Yamada says. “That’s one of the benefits we have now: We can run longer simulations because we can run more time steps within a fixed wall-clock time.”
Finally, the new capability makes it easier to run suites of simulations, changing conditions like temperature, cooling rate and surface quality in each. That’s important because nucleation is random, Yamada says: “Sometimes nanodroplets never freeze and sometimes they freeze within the time you’re looking at them.” Multiple simulations allow researchers to chart a distribution of freezing time under varying conditions.
Even with these capabilities, a million-molecule simulation is still a tiny fraction of the smallest real drops. And molecular dynamics simulations on the scale of a full wind-turbine blade would take all the world’s computers hundreds of years – or more, Yamada says.
This is the freezing project’s second ASCR challenge allocation on an Oak Ridge machine. The first, also for 40 million hours, studied freezing on six different surfaces at varying temperatures. The study gave scientists an idea of how varying parameters affect freezing behavior.
The latest work focuses on further accelerating the simulations and on a curious phenomenon: Just as a supercooled water droplet is at the tipping point to freeze, its temperature spikes, releasing latent heat that must be carried away from the advancing ice front. The researchers want to understand heat removal as an additional parameter affecting the freezing process.
It’s been an engaging project, Yamada says. The public is excited because it could lead to more efficient clean energy. Physicists are intrigued because there’s a lot they don’t know about nucleation. And Oak Ridge computer scientists like that it makes good use of their machine.
“It covers the bases of what I think the supercomputing people are really interested in: seeing their apparatus utilized to solve a meaningful, interesting problem.”

